by Buck Institute
April 5, 2018 . Press Release
Largest “pan-cancer” genomics study spurs future efforts to enroll patients in specialized “basket” clinical trials
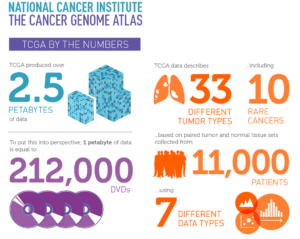
Groundbreaking TCGA genomic analysis of 33 different tumor types from more than 10,000 patients sets the stage for improved cancer classification and new treatment approaches.
NOVATO, Calif. — The final output from the largest-ever cancer genomics study reveals new possibilities for immune-based and other novel cancer therapeutics and provides a push for clinicians to obtain and utilize comprehensive genomic information to enroll their patients in specialized “basket” or “umbrella” clinical trials. Results from The Cancer Genome Atlas (TCGA) Research Network are highlighted in 27 studies published this week in Cell, Cancer Cell, Cell Reports, and Immunity.
The TCGA Research Network, which over the past decade has involved several hundred researchers from the United States and abroad, painstakingly analyzed the DNA, RNA, and protein of 33 tumor types from more than 10,000 patients. Prior to study accrual, the tumors were classically identified by their anatomic site of origin, such as breast, kidney, lung, and so forth. Utilizing four to six different state-of-the-art technology platforms to assay all the tumor samples, researchers found that, based on their cellular and genetic makeup and independent of their anatomic site of origin, all 33 tumor types could be reclassified into 28 different molecular types or “clusters” and that nearly two-thirds of these clusters were considered heterogeneous as they contained up to 25 different histological tumor types that traditionally would all be treated differently. These molecular analyses and clustering results, now also linked to multiple clinical outcome endpoints, are available to clinicians and researchers worldwide.
“This comprehensive body of final TCGA Pan-Cancer Atlas analyses will provide a new foundation for future cancer research efforts and clinical trials. It will also incentivize clinical oncologists to get newly diagnosed and recurrent tumors genomically characterized and encourage them to help their patients become enrolled in specialized precision-medicine basket clinical trials evaluating promising new targeted therapies against molecularly similar tumors, despite different histologies and anatomic sites of origin,” said Christopher Benz, MD, professor of cancer and developmental therapeutics at the Buck Institute for Research on Aging and a clinical oncologist at the University of California, San Francisco. “Patients will have the best shot at successful treatment if their tumors can first be classified according to their genomic and molecular makeup,” he continued. Benz has been involved in the TCGA Research Network since its inception in 2005 and is a senior co-author on several of the papers being published in Cell.
New data boosts the promise of immunotherapies
Benz says the new TCGA data holds particular promise for expanding treatments designed to enlist the immune system to beat cancer, including approved immunotherapies that are now showing extremely promising results against a limited number of classical cancer types. Remarkably, the study shows that one of the most diverse of the observed 28 molecular clusters was composed of 25 different classical tumor types and exhibited very strong features linked to activation of the patient’s immune response. “This finding supports the growing notion that specific immunotherapies approved by the FDA for one cancer type would likely benefit patients with various other cancer types, if these other types could be molecularly identified,” Benz said. “These latest Pan-Cancer Atlas results will likely be of great interest to pharmaceutical companies [that are] scrambling to develop new cancer therapies based on immune function or interested in repurposing some of their already-approved agents based on this new molecular classification.”
Repurposing drugs used for other diseases
Drugs approved for other diseases could also be effective against some of the newly classified cancer types. “A couple of our newly defined cancer clusters also show activation of a molecular pathway (JAK/Stat) that’s commonly upregulated in rheumatoid arthritis,” said Christina Yau, PhD, a senior scientist in the Benz lab and new UCSF faculty member who provided bioinformatics expertise for much of TCGA’s work over the past decade. “This may provide the molecular rationale to explore repurposing of drugs used to treat that nonmalignant chronic disease, as a novel treatment strategy.”
Even though TCGA is done — the database won’t be added to or changed — this same kind of comprehensive and collaborative multiplatform genomic analysis continues nationwide under new National Cancer Institute sponsorship. As with their TCGA efforts, Benz and Yau continue to team up with University of California, Santa Cruz, principal investigator Josh Stuart and his colleagues as part of their bioinformatic analysis center for the newly established Genome Data Analysis Network. Among other challenges, the center is tasked with determining clinically measurable biomarkers that would make it easier and more cost effective to identify a priori those same tumor molecular subsets identified by the TCGA Research Network’s multiplatform analysis. Benz attributes the innovative and highly effective bioinformatic algorithms developed by his UCSC partners for much of the shared success of their TCGA Buck-UCSC Genome Data Analysis Center (GDAC) over the past decade.
“It’s time to rewrite the textbooks on cancer, and it’s time to break down the silos in clinical oncology that make it difficult for patients to take advantage of this paradigm shift in cancer classification,” said Benz. “This body of TCGA work is crucial for the success of our nation’s ‘Cancer Moonshot’ initiative, and all of us who have been involved in the TCGA are excited to make it available to the worldwide community of cancer researchers.”
This research was covered widely in the media, including on the BBC, in The Telegraph, and The Marin Independent Journal.
Citation: Hoadley, K. A., Yau, C., Hinoue, T., Wolf, D. M., . . . Benz, C. C., and Laird, P. W. (2018, Apr. 5). Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell, 173(2), 291–304. DOI: 10.1016/j.cell.2018.03.022
Citation: Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., . . . and Hu, H. (2018, Apr. 5). An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell, 173(2), 400–416. DOI: 10.1016/j.cell.2018.02.052
Science is showing that while chronological aging is inevitable, biological aging is malleable. There's a part of it that you can fight, and we are getting closer and closer to winning that fight.
Eric Verdin, MD, Buck Institute President and CEO